During development, cells must contend with the physical forces exerted by replication and transcription to successfully differentiate. Interpreting genetic and genomic data thus requires an understanding of DNA as a tangible, dynamic object with concrete physical properties.
I am interested how these properties connect to the development and evolution of cell states, especially in the human nervous system. I study genome architecture, gene composition, and physical genomic forces towards building a complete model of mammalian gene regulation.
My research experience spans topics in gene regulation, neuroscience, and drug development. Prior to graduate school, I helped to design and test landmark transcriptome and epigenome engineering tools in mammals, enabling optical control and genome-scale screening. During my Ph.D., I studied the development of the human cerebral cortex and explored general physical rules that govern gene regulation. Finally, I transitioned to a role at a startup biotechnology company, where I established and led a machine learning R&D group that has made substantial published contributions to topics in spatial biology data analysis.
1. Curiosity First: leave time to think – follow rabbit holes – take risks
2. Active Bias: be hands-on when learning – design hypothesis-driven experiments
3. More to Learn: ask questions – good experiments > good results
4. Collectivism: share opportunities – lift others up – promote flourishing
I. The Physical Genome
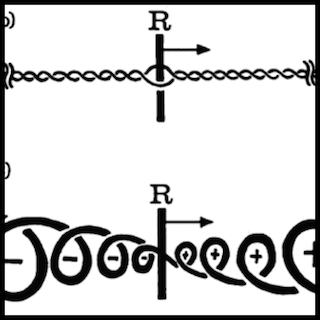
Architecture, constraints, and functional consequences
II. Neural Evolution and Development
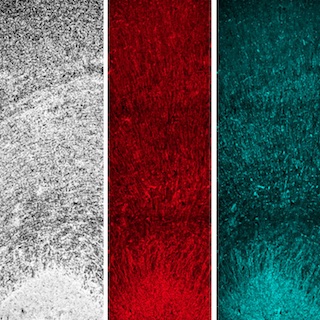
Unique genomic and molecular features of the human nervous system and their implications in disease
III. Patient Centric Drug Development

Connecting molecular profiling to real-world patient experiences